Spanner is a structural homology modeling pipeline that threads a query amino-acid sequence onto a template protein structure. Spanner is unique in that it handles gaps by spanning the region of interest using fragments of known structures.
To create a model, you must provide a template structure, as well as an alignment of the query sequence you wish to model onto the template sequence. Spanner will replace mismatched residues, and fill any gaps caused by insertions or deletions.
For users that are unable to create an alignment a method for building a model starting only from sequence is also available. During this process a template search is conducted and an alignment is built dynamically using FORTE before being passed through to the main part of the pipeline.
Spanner consists of several modules written in the Go programming language. For Spanner jobs which build a model only from sequence, the first step is a search of the PDB for possible templates using BLAST+.1 These possible templates are then aligned and scored with FORTE.2
The next step involves defining the start and end points of fragments corresponding to insertions or deletions. The start and end points are referred to as anchors because they must be equivalent in both the template and any candidate fragment. The margin parameter determines how far from the edge of a gap the fragment begins or ends. For example a margin of 0 would mean that the anchors begins at the very edge of a gap. This is usually not a good idea, and the default margin is set to 1.
A representative set of protein chains was prepared using CD-HIT at 100% sequence identity.3 All continuous fragments were then extracted from this set of chains and stored in a relational database, indexed by the internal coordinates of the fragment endpoints. A separate database is prepared for each fragment length. Currently, fragments of length 8-40, including the 8 anchor residues, are stored in the database.
For a given fragment, a fragment index is generated from the template anchor residues. A tolerance in the fit to the anchor residues is used to specify a range of index values. The index range is used to generate a database query for the appropriate fragment length, and all fragments satisfying the range of indices are returned. Since the number of returned fragments is sensitive to the tolerance in the fit to the anchor residues, the retrieval step starts with a small value of 0.5 Å, and incrementally increases the tolerance until the required number of fragments (1000) or a maximum tolerance (2.5 Å) is reached.
The fragments returned from the DB are then sorted by a simple score that is a function only of the primary and secondary structure similarities:
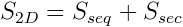
where S seq is proportional to a log-odds sequence substitution matrix score derived from a large number of structure alignments4 and S sec is proportional to a secondary structure substitution matrix score.5 The returned candidate fragments are then re-scored using a more sensitive function that takes structure into account and is given by:
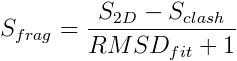
where S clash is a weighted sum of clashes between the fragment and the rest of the template structure excluding residues that are to be replaced by the fragment, and RMSD fit is given by the root-mean square deviation of C α atoms in the fitted anchor residues. The fragment with the top score is then inserted into the template structure. When necessary, side-chains are re-modeled with SCWRL4.6
We are always looking to improve Spanner based on user feedback. Please contact us with any bug reports or feedback you may have.